Introduction to Data Analysis with Python
Erick Martins Ratamero
Research Fellow
- These slides: http://tiny.cc/camdupython2 (redirects to https://erickmartins.github.io/DataAnalysisPython.html)
- Tidy data
- Some relevant libraries
- String manipulation
- File input/output
- Plotting
- If we have time: introduction to pandas and numpy
"Happy families are all alike; every unhappy family is unhappy in its own way"
Leo Tolstoy
We will discuss a standardised way to link the structure of a dataset to its semantics.
(i.e. we will relate the physical layout of data to its meaning in a predictable way.)
import pandas as pd
test = pd.read_csv("test.csv")
test
name condition1 condition2
1 Alice NA 8
2 Bob 4 2
3 Carol 6 9
import pandas as pd
test2 = pd.read_csv("test2.csv")
test2
condition Alice Bob Carol
1 1 NA 4 6
2 2 8 2 9
Data is the same, but layout is different - no easy way of knowing it is the same data!
A dataset is a collection of values.
A value belongs to a variable and an observation.
A variable contains all values that measure the same attribute (height, duration, frequency).
An observation contains all values measured on the same "experiment".
import pandas as pd
test_tidy = pd.read_csv("test_tidy.csv")
test_tidy
name condition n
1 Alice 1 NA
2 Alice 2 8
3 Bob 1 4
4 Bob 2 2
5 Carol 1 6
6 Carol 2 9
Tidy data depends on your experimental design.
Each variable should be a column.
Each observation should be a row.
Each type of observation (demographic, medical, meteorological, etc.) should be a table.
- Column headers as values instead of variable names
- Multiple variables in the same column
- Variables in rows and columns
- Multiple types of obervations in the same table
- Same type of observation in different tables
religion `<$10k` `$10-20k` `$20-30k` `$30-40k` `$40-50k` `$50-75k`
1 Agnostic 27 34 60 81 76 137
2 Atheist 12 27 37 52 35 70
3 Buddhist 27 21 30 34 33 58
4 Catholic 418 617 732 670 638 1116
5 Don’t k… 15 14 15 11 10 35
6 Evangel… 575 869 1064 982 881 1486
7 Hindu 1 9 7 9 11 34
8 Histori… 228 244 236 238 197 223
9 Jehovah… 20 27 24 24 21 30
10 Jewish 19 19 25 25 30 95
iso2 year m04 m514 m014 m1524 m2534 m3544 m4554 m5564 m65 mu
1 AD 1989 NA NA NA NA NA NA NA NA NA NA
2 AD 1990 NA NA NA NA NA NA NA NA NA NA
3 AD 1991 NA NA NA NA NA NA NA NA NA NA
4 AD 1992 NA NA NA NA NA NA NA NA NA NA
5 AD 1993 NA NA NA NA NA NA NA NA NA NA
6 AD 1994 NA NA NA NA NA NA NA NA NA NA
7 AD 1996 NA NA 0 0 0 4 1 0 0 NA
8 AD 1997 NA NA 0 0 1 2 2 1 6 NA
9 AD 1998 NA NA 0 0 0 1 0 0 0 NA
10 AD 1999 NA NA 0 0 0 1 1 0 0 NA
id year month element d1 d2 d3 d4 d5 d6 d7
1 MX17… 2010 1 tmax NA NA NA NA NA NA NA
2 MX17… 2010 1 tmin NA NA NA NA NA NA NA
3 MX17… 2010 2 tmax NA 27.3 24.1 NA NA NA NA
4 MX17… 2010 2 tmin NA 14.4 14.4 NA NA NA NA
5 MX17… 2010 3 tmax NA NA NA NA 32.1 NA NA
6 MX17… 2010 3 tmin NA NA NA NA 14.2 NA NA
7 MX17… 2010 4 tmax NA NA NA NA NA NA NA
8 MX17… 2010 4 tmin NA NA NA NA NA NA NA
9 MX17… 2010 5 tmax NA NA NA NA NA NA NA
10 MX17… 2010 5 tmin NA NA NA NA NA NA NA
- numpy: NUMerical PYthon. Has multidimensional arrays, basic linear algebra, Fourier transforms, random number generation...
- scipy: SCIentific PYthon. Higher-level science and engineering modules (optimization, e.g.).
- matplotlib: Plotting library. Histograms, line plots, heat plots...
- pandas: Structured data manipulation and operations. Data scientists love this one.
- seaborn: Also does plotting, but it looks nice compared to matplotlib.
- os: Opeating system stuff and file operations.
We've talked a little bit about strings before.
What if we want a string containing a quote?
string = 'That is Erick's presentationFix: use double quotes!
string = "That is Erick's presentation"What if we need double quotes in the string?
\'Single quote\"Double quote\tTab\nNew line (or line break)\\backslash
>>> print("Hello there!\nHow are you?\nI\'m doing fine.")
Hello there!
How are you?
I'm doing fine.
Strings use indexing the same way lists do. It's just a list of characters! The space and exclamation points count, so 'Hello world!' has 12 characters (from 0 to 11).
>>> test = 'Hello world!'
>>> test[0]
'H'
>>> test[4]
'o'
>>> test[-1]
'!'
>>> test[0:5]
'Hello'
>>> test[:5]
'Hello'
>>> test[6:]
'world!'The in and not in operators work for strings as well as lists (have I mentioned that strings are just lists of characters?). It will return True or False.
>>> 'Hello' in 'Hello world!'
True
>>> 'Hello' in 'Hello'
True
>>> 'HELLO' in 'Hello world!'
False
>>> '' in 'test'
True
>>> 'cats' not in 'cats and dogs'
FalseThe startswith() and endswith() methods return True if the string they are called on begins or (respectively) ends with the string passed to the method.
>>> 'Hello world!'.startswith('Hello')
True
>>> 'Hello world!'.endswith('world!')
True
>>> 'abc123'.startswith('abcdef')
False
>>> 'abc123'.endswith('12')
False
>>> 'Hello world!'.startswith('Hello world!')
True
>>> 'Hello world!'.endswith('Hello world!')
True
The join() and split() do what you expect them to do. split() can take a string argument and will split the string upon the occurrence of this argument. It is especially useful when dealing with CSV files, as we will see later.
>>> ', '.join(['cats', 'rats', 'bats'])
'cats, rats, bats'
>>> ' '.join(['My', 'name', 'is', 'Simon'])
'My name is Simon'
>>> 'ABC'.join(['My', 'name', 'is', 'Simon'])
'MyABCnameABCisABCSimon'
>>> 'My name is Simon'.split()
['My', 'name', 'is', 'Simon']
>>> 'MyABCnameABCisABCSimon'.split('ABC')
['My', 'name', 'is', 'Simon']
>>> 'My name is Simon'.split('m')
['My na', 'e is Si', 'on']
We can use strip() and its friends to remove whitespace characters (space, tab, newline) from the ends of a string. It returns a new string without the whitespaces.
>>> spam = ' Hello World '
>>> spam.strip()
'Hello World'
>>> spam.lstrip()
'Hello World '
>>> spam.rstrip()
' Hello World'
Alternatively, you can tell strip() which characters you want to remove.
>>> spam = 'SpamSpamBaconSpamEggsSpamSpam'
>>> spam.strip('ampS')
'BaconSpamEggs'
- Write a Python program to calculate the length of a string.
- Write a Python program to add 'ing' at the end of a given string (length should be at least 3). If the given string already ends with 'ing' then add 'ly' instead. If the string length of the given string is less than 3, leave it unchanged. ('abc' returns 'abcing', 'string' returns 'stringly', etc etc)
Sample solution 1
def string_length(str1):
count = 0
for char in str1:
count += 1
return count
print(string_length('test'))Sample solution 2
def add_string(str1):
length = len(str1)
if length > 2:
if str1[-3:] == 'ing':
str1 += 'ly'
else:
str1 += 'ing'
return str1
print(add_string('ab'))
print(add_string('abc'))
print(add_string('string'))- Every file contains three key pieces of information: path, filename and extension.
- path: indicates in which folder the file exists. Can be something like
C:\Users\User1\Desktop\on Windows and/home/User1/Desktopon Mac/Linux. - filename: the actual name of the file. Can be essentially anything you want except for some special characters.
- extension: indicates the file type. Common extensions are
.doc, .xls, .txtand so on. They are often hidden in Windows.
As you can see from the examples, Windows has a different way of indicating folders compared to Mac/Linux, including some weird backslashes.
If you want your code to run in any system, it needs to be aware of which system it is! os.path.join() is your best friend.
>>> import os
>>> os.path.join('usr', 'bin', 'spam')
'usr\\bin\\spam'Every program has a current working directory - every path that doesn't start with the root folder (C:\ on Windows, / on Mac/Linux) is presumed to be inside the current working directory.
>>> import os
>>> os.getcwd()
'C:\\Python34'
>>> os.chdir('C:\\Windows\\System32')
>>> os.getcwd()
'C:\\Windows\\System32'File paths can be absolute (starts with root directory) or relative (from the current working directory).

Some additional os functions that can be useful:
>>> os.path.abspath('.\\Scripts')
'C:\\Python34\\Scripts'
>>> os.path.relpath('C:\\Windows', 'C:\\spam\\eggs')
'..\\..\\Windows'
>>> os.getcwd()
'C:\\Python34'
>>> path = 'C:\\Windows\\System32\\calc.exe'
>>> os.path.basename(path)
'calc.exe'
>>> os.path.dirname(path)
'C:\\Windows\\System32'
>>> os.path.split(path)
('C:\\Windows\\System32', 'calc.exe')
>>> os.path.exists('C:\\Windows')
True
>>> os.path.exists('C:\\some_made_up_folder')
False
>>> os.path.isdir('C:\\Windows\\System32')
True
>>> os.path.isfile('C:\\Windows\\System32')
False
>>> os.path.isdir('C:\\Windows\\System32\\calc.exe')
False
>>> os.path.isfile('C:\\Windows\\System32\\calc.exe')
True
For finding everything that is inside a folder, use os.listdir(path):
>>> os.listdir('C:\\Windows\\System32')
['0409', '12520437.cpx', '12520850.cpx', '5U877.ax', 'aaclient.dll',
--snip--
'xwtpdui.dll', 'xwtpw32.dll', 'zh-CN', 'zh-HK', 'zh-TW', 'zipfldr.dll']If you want to recursively go into every subfolder, you will need something like this:
>>> import os
>>> path = '/home/erick/Desktop'
>>> for root,dirs,files in os.walk('.'):
... print("current directory is "+root)
... print("immediate subdirectories are "+' '.join(dirs))
... print("files in this subfolder are "+' '.join(files))
...
current directory is .
immediate subdirectories are js img-ia css .git plugin test img img-py lib material
files in this subfolder are test.md bower.json handout.png DataAnalysisPython.html package.json LICENSE ImageAnalysisWithFiji.html CONTRIBUTING.md index.html README.md Gruntfile.js README_REVEAL.md IntroPython.html ImageAnalysis.html
current directory is ./js
(...)
To read or write files, you first need to open them. Try saving the file hello.txt file on your home folder and then opening it:
>>> helloFile = open('C:\\Users\\your_home_folder\\hello.txt')Or if you're Mac/Linux
>>> helloFile = open('/Users/your_home_folder/hello.txt')This will open the file in read-only mode. If you want to open the file in write mode:
>>> helloFile = open('/Users/your_home_folder/hello.txt', 'w')Two ways of reading the contents of a file (try it yourself with the sonnet29.txt file):
>>> f = open('/Users/your_home_folder/sonnet29.txt')
>>> f.read()
"When, in disgrace with fortune and men's eyes,\nI all alone beweep my outcast state,\nAnd trouble deaf heaven with my bootless cries,\nAnd look upon myself and curse my fate,"
>>> f2 = open('material/sonnet29.txt')
>>> f2.readlines()
["When, in disgrace with fortune and men's eyes,\n", 'I all alone beweep my outcast state,\n', 'And trouble deaf heaven with my bootless cries,\n', 'And look upon myself and curse my fate,']
Now let's try writing to a file. Remember: we need to use 'w' when opening it. After that, we just need to use the write method:
>>> myFile = open('test.txt', 'w')
>>> myFile.write('Hello world!\n')
>>> myFile.close()
>>> myFile = open('test.txt', 'a')
>>> myFile.write('This is a nice extra message!')
>>> myFile.close()
>>> myFile = open('test.txt')
>>> content = myFile.read()
>>> myFile.close()
>>> print(content)
Hello world!
This is a nice extra message!
- Write a Python program to print the first 5 lines of the cereal.csv file.
- Write a Python program to print the first 5 columns of the cereal.csv file.
- Write a Python program to print all lines of the cereal.csv file where calories >= 120.
Sample solution 1
import os
os.chdir('Whatever directory your cereal.csv file is in')
f = open('cereal.csv')
lines = f.readlines()
for i in range(5):
print(lines[i])
f.close()Sample solution 2
import os
os.chdir('Whatever directory your cereal.csv file is in')
f = open('cereal.csv')
lines = f.readlines()
for line in lines:
line_split = line.split(',')
print(','.join(line_split[:5])+'\n')
f.close()Sample solution 3
import os
os.chdir('Whatever directory your cereal.csv file is in')
f = open('cereal.csv')
lines = f.readlines()[1:]
for line in lines:
line_split = line.split(',')
cal = int(line_split[3])
if (cal >= 120):
print(line)
f.close()Our best friend in this section will be matplotlib.pyplot
import matplotlib.pyplot as plt
Let's start with an advanced example and then move back to the basics.
import matplotlib.pyplot as plt
import numpy as np
rng = np.arange(50)
rnd = np.random.randint(0, 10, size=(3, rng.size))
yrs = 1950 + rng
fig, ax = plt.subplots(figsize=(5, 3))
ax.stackplot(yrs, rng + rnd, labels=['Eastasia', 'Eurasia', 'Oceania'])
ax.set_title('Combined debt growth over time')
ax.legend(loc='upper left')
ax.set_ylabel('Total debt')
ax.set_xlim(left=yrs[0], right=yrs[-1])
fig.tight_layout()
plt.show()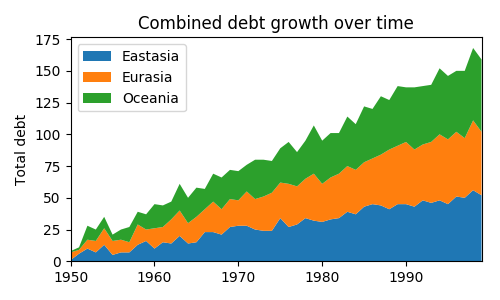
A basic example:
import matplotlib.pyplot as plt
plt.plot([1,2,3,4])
plt.ylabel('some numbers')
plt.show()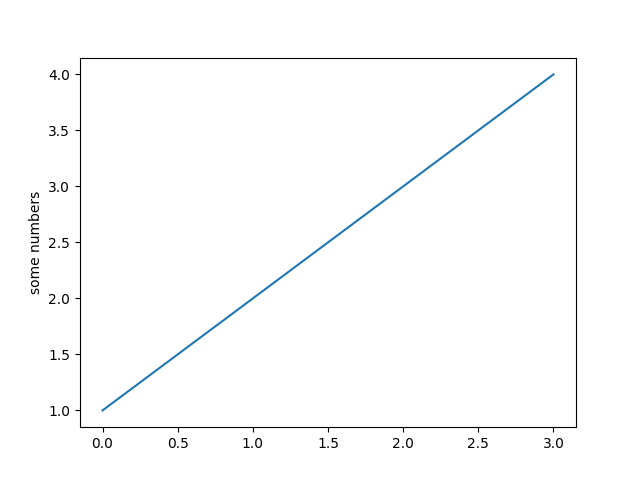
Changing default format ('b-') to red circles ('ro'):
import matplotlib.pyplot as plt
plt.plot([1,2,3,4], [1,4,9,16], 'ro')
plt.axis([0, 6, 0, 20])
plt.show()
We can have multiple plots in the same axes.
import numpy as np
import matplotlib.pyplot as plt
t = np.arange(0., 5., 0.2)
# red dashes, blue squares and green triangles
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
plt.show()
- Plot the following dataset in Python. Use a blue dashed line. Don't forget to add a title and axis labels!
| Time (decade): | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
| CO2 concentration (ppm): | 250 | 265 | 272 | 260 | 300 | 320 | 389 |
- Repeat the multiple plots in the same figure as a few slides ago, but using the following properties:
- y = t: teal, dotted line
- y = t^2: Yellow, dashed line
- y = t^3: Magenta circles
A few sites that might be useful:
Sample solution 1
import matplotlib.pyplot as plt
times = range(7)
co2 = [250, 265, 272, 260, 300, 320, 389]
#plt.plot(times, co2)
plt.plot(times, co2, 'b--')
plt.title("Concentration of CO2 versus time")
plt.ylabel("[CO2]")
plt.xlabel("Time (decade)")
plt.show()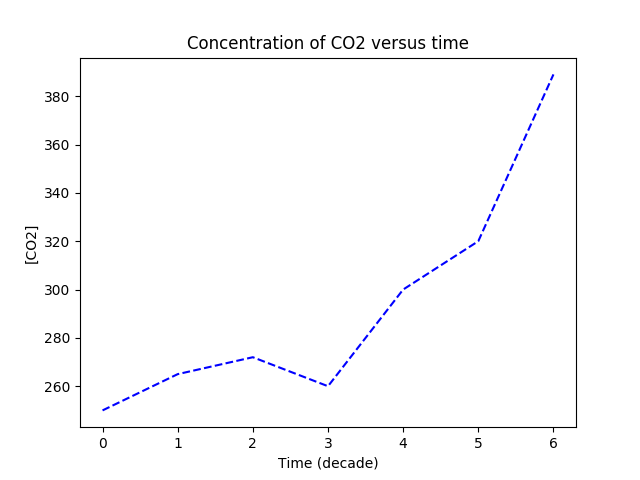
Sample solution 2
import numpy as np
import matplotlib.pyplot as plt
t = np.arange(0., 5., 0.2)
plt.plot(t, t, color='teal', linestyle=':')
plt.plot(t, t**2, 'y--', t, t**3, 'mo')
plt.show()
Pandas is a data analysis/manipulation library. It operates on top of the numerical library numpy, so we will start by having a look at it.
>>> import numpy as np
>>> np.zeros(10)
array([0., 0., 0., 0., 0., 0., 0., 0., 0., 0.])
>>> np.full((3,5),1.23)
array([[1.23, 1.23, 1.23, 1.23, 1.23],
[1.23, 1.23, 1.23, 1.23, 1.23],
[1.23, 1.23, 1.23, 1.23, 1.23]])
>>> np.arange(0, 20, 2)
array([ 0, 2, 4, 6, 8, 10, 12, 14, 16, 18])
>>> np.linspace(0, 1, 5)
array([0. , 0.25, 0.5 , 0.75, 1. ])
>>> np.random.randint(10, size=6)
array([0, 4, 1, 3, 9, 1])
>>> np.random.randint(10, size=(3,4))
array([[0, 3, 5, 7],
[3, 4, 7, 2],
[5, 2, 1, 0]])
>>> np.random.randint(10, size=(3,4,5))
array([[[5, 3, 2, 5, 6],
[2, 6, 0, 2, 3],
[3, 7, 7, 7, 5],
[5, 1, 1, 5, 9]],
[[5, 4, 0, 0, 1],
[6, 4, 2, 6, 5],
[6, 1, 7, 4, 8],
[2, 1, 7, 9, 4]],
[[7, 1, 9, 2, 7],
[5, 8, 3, 3, 0],
[6, 5, 9, 1, 4],
[0, 4, 7, 8, 4]]])
Indexing and slicing work in the same way as they do for lists.
>>> x1 = np.array([4, 3, 4, 4, 8, 4])
>>> x1[0]
4
>>> x1[-2]
8
>>> x2 = np.array([[3, 7, 5, 5],
... [0, 1, 5, 9],
... [3, 0, 5, 0]])
>>> x2[2,3]
0
>>> x2[2,-1]
0
>>> x = np.arange(10)
>>> x[:5]
array([0, 1, 2, 3, 4])
>>> x[4:7]
array([4, 5, 6])
Numpy arrays can be concatenated and split in many different ways.
>>> x = np.array([1, 2, 3])
>>> y = np.array([3, 2, 1])
>>> z = [21,21,21]
>>> np.concatenate([x, y,z])
array([ 1, 2, 3, 3, 2, 1, 21, 21, 21])
>>> x = np.array([3,4,5])
>>> grid = np.array([[1,2,3],[17,18,19]])
>>> np.vstack([x,grid])
array([[ 3, 4, 5],
[ 1, 2, 3],
[17, 18, 19]])
>>> z = np.array([[9],[9]])
>>> np.hstack([grid,z])
array([[ 1, 2, 3, 9],
[17, 18, 19, 9]])
>>> grid = np.arange(16).reshape((4,4))
>>> grid
array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11],
[12, 13, 14, 15]])
>>> upper,lower = np.vsplit(grid,[2])
>>> print (upper, lower)
(array([[0, 1, 2, 3],
[4, 5, 6, 7]]), array([[ 8, 9, 10, 11],
[12, 13, 14, 15]]))
The main Pandas structure we will be interested in is the DataFrame.
import pandas as pd
data = pd.DataFrame({'Country': ['Russia','Colombia','Chile','Equador','Nigeria'],
'Rank':[121,40,100,130,11]})
print(data)
Country Rank
0 Russia 121
1 Colombia 40
2 Chile 100
3 Equador 130
4 Nigeria 11
A quick way of having a summary of the statistics your data set is using the describe() method.
data.describe()
Rank
count 5.000000
mean 80.400000
std 52.300096
min 11.000000
25% 40.000000
50% 100.000000
75% 121.000000
max 130.000000
Of course, that will only give you information about the numeric fields of your dataset. If you want more general information, use info().
>>> data.info()
RangeIndex: 5 entries, 0 to 4
Data columns (total 2 columns):
Country 5 non-null object
Rank 5 non-null int64
dtypes: int64(1), object(1)
memory usage: 160.0+ bytes
We can, of course, sort the data set.
>>> data.sort_values(by=['Rank'],ascending=True,inplace=False)
Country Rank
4 Nigeria 11
1 Colombia 40
2 Chile 100
0 Russia 121
3 Equador 130
You can sort by more than one column.
>>> data = pd.DataFrame({'k1':['one']*3 + ['two']*4 + ['one']*2, 'k2':[2,1,3,3,3,4,4,2,5]})
>>> data
k1 k2
0 one 2
1 one 1
2 one 3
3 two 3
4 two 3
5 two 4
6 two 4
7 one 2
8 one 5
>>> data.sort_values(by=['k1','k2'],ascending=[True,False],inplace=False)
k1 k2
8 one 5
2 one 3
0 one 2
7 one 2
1 one 1
5 two 4
6 two 4
3 two 3
4 two 3
Removing duplicates is very easy!
>>> data.drop_duplicates()
k1 k2
0 one 2
1 one 1
2 one 3
3 two 3
5 two 4
8 one 5
This is how you create a new variable based on a combination of the existing ones:
>>> data.assign(new_variable = data['k2']*20)
k1 k2 new_variable
0 one 2 40
1 one 1 20
2 one 3 60
3 two 3 60
4 two 3 60
5 two 4 80
6 two 4 80
7 one 2 40
8 one 5 100
Now, let's try binning some data.
data = pd.DataFrame({'names':['Alice','Bob','Charlie','Dan','Eve','Frank','Grace','Heidi','Judy','Michael','Oscar','Pat'], 'ages':[20, 22, 25, 27, 21, 23, 37, 31, 61, 45, 41, 32]})
>>> data
names ages
0 Alice 20
1 Bob 22
2 Charlie 25
3 Dan 27
4 Eve 21
5 Frank 23
6 Grace 37
7 Heidi 31
8 Judy 61
9 Michael 45
10 Oscar 41
11 Pat 32
>>> bins = [18, 25, 35, 60, 100]
>>> brackets = pd.cut(data['ages'],bins)
>>> brackets
0 (18, 25]
1 (18, 25]
2 (18, 25]
3 (25, 35]
4 (18, 25]
5 (18, 25]
6 (35, 60]
7 (25, 35]
8 (60, 100]
9 (35, 60]
10 (35, 60]
11 (25, 35]
Name: ages, dtype: category
Categories (4, interval[int64]): [(18, 25] < (25, 35] < (35, 60] < (60, 100]]
>>> pd.value_counts(brackets)
(18, 25] 5
(35, 60] 3
(25, 35] 3
(60, 100] 1
Name: ages, dtype: int64
A very useful operation for datasets is grouping data and creating summaries.
>>> df = pd.DataFrame({'key1' : ['a', 'a', 'b', 'b', 'a'],
'key2' : ['one', 'two', 'one', 'two', 'one'],
'data1' : np.random.randn(5),
'data2' : np.random.randn(5)})
>>> df
key1 key2 data1 data2
0 a one -0.369708 -1.215059
1 a two -0.341208 -0.884716
2 b one -0.672269 0.149367
3 b two -1.468556 1.386700
4 a one -0.366804 -0.634380
>>> grouped = df['data1'].groupby(df['key1'])
>>> grouped.mean()
key1
a -0.359240
b -1.070413
Name: data1, dtype: float64
There are many, many ways of accessing specific subsets of our data set.
>>> dates = pd.date_range('20130101',periods=6)
>>> df = pd.DataFrame(np.random.randn(6,4),index=dates,columns=list('ABCD'))
>>> df
A B C D
2013-01-01 0.482837 0.005617 1.011802 -0.817221
2013-01-02 -1.351376 0.552897 0.503408 -0.431504
2013-01-03 -0.244983 -0.470076 -0.489829 -0.763309
2013-01-04 0.019689 -1.759921 0.740204 0.763700
2013-01-05 0.607637 1.679538 -0.599785 -0.365752
2013-01-06 1.371928 -1.148383 -1.586073 0.784124
>>> df[:3]
A B C D
2013-01-01 0.482837 0.005617 1.011802 -0.817221
2013-01-02 -1.351376 0.552897 0.503408 -0.431504
2013-01-03 -0.244983 -0.470076 -0.489829 -0.763309
>>> df['20130101':'20130104']
A B C D
2013-01-01 0.482837 0.005617 1.011802 -0.817221
2013-01-02 -1.351376 0.552897 0.503408 -0.431504
2013-01-03 -0.244983 -0.470076 -0.489829 -0.763309
2013-01-04 0.019689 -1.759921 0.740204 0.763700
There are many, many ways of accessing specific subsets of our data set.
>>> df.loc[:,['A','B']]
A B
2013-01-01 0.482837 0.005617
2013-01-02 -1.351376 0.552897
2013-01-03 -0.244983 -0.470076
2013-01-04 0.019689 -1.759921
2013-01-05 0.607637 1.679538
2013-01-06 1.371928 -1.148383
>>> df.loc['20130102':'20130103',['A','B']]
A B
2013-01-02 -1.351376 0.552897
2013-01-03 -0.244983 -0.470076
>>> df[df.A > 1]
A B C D
2013-01-06 1.371928 -1.148383 -1.586073 0.784124
>>> df.query('A > C')
A B C D
2013-01-03 -0.244983 -0.470076 -0.489829 -0.763309
2013-01-05 0.607637 1.679538 -0.599785 -0.365752
2013-01-06 1.371928 -1.148383 -1.586073 0.784124
Pandas can do plotting, too!
>>> df = pd.DataFrame({
'name':['john','mary','peter','jeff','bill','lisa','jose'],
'age':[23,78,22,19,45,33,20],
'gender':['M','F','M','M','M','F','M'],
'state':['california','dc','california','dc','california','texas','texas'],
'num_children':[2,0,0,3,2,1,4],
'num_pets':[5,1,0,5,2,2,3]
})
>>> import matplotlib.pyplot as plt
>>> df.plot(kind='scatter',x='num_children',y='num_pets',color='red')
<matplotlib.axes._subplots.AxesSubplot object at 0x7f56722e22e8>
>>> plt.show()
>>> df.plot(kind='bar',x='name',y='age')
<matplotlib.axes._subplots.AxesSubplot object at 0x7f56722e22e8>
>>> plt.show()
>>> fig, ax = plt.subplots()
>>> df.plot(kind='line',x='name',y='num_children',ax=ax)
<matplotlib.axes._subplots.AxesSubplot object at 0x7f56622145c0>
>>> df.plot(kind='line',x='name',y='num_pets', color='red', ax=ax)
<matplotlib.axes._subplots.AxesSubplot object at 0x7f56622145c0>
>>> plt.show()
CSV files are pandas' best friend. Try this with cereal.csv:
>>> import os
>>> os.chdir('Whatever directory you\'ve saved cereal.csv in')
>>> df = pd.read_csv('cereal.csv')
>>> df.head(5)
name mfr type ... weight cups rating
0 100% Bran N C ... 1.0 0.33 68.402973
1 100% Natural Bran Q C ... 1.0 1.00 33.983679
2 All-Bran K C ... 1.0 0.33 59.425505
3 All-Bran with Extra Fiber K C ... 1.0 0.50 93.704912
4 Almond Delight R C ... 1.0 0.75 34.384843
[5 rows x 16 columns]
You can also easily save a dataframe as a CSV file:
>>> df.to_csv('cereal_copy.csv')
Do you see any difference between the copies?
- Write a Python program to plot all the data in fdata.csv.
- Write a Python program to create a 5x5 numpy array with 1 on the border and 0 inside.
- Write a Python program to create a 5x5 array with random values and find the minimum and maximum values.
- Write a Python program to iterate over rows in a DataFrame.
A few sites that might be useful:
Sample solution 1
import matplotlib.pyplot as plt
import pandas as pd
import os
os.chdir('Whatever directory you\'ve saved fdata.csv in')
df = pd.read_csv('fdata.csv')
df.plot(x="Date",y=["Open", "High", "Low", "Close"])
plt.show()
Sample solution 2
import numpy as np
x = np.ones((5,5))
x[1:-1,1:-1] = 0
print(x)
[[1. 1. 1. 1. 1.]
[1. 0. 0. 0. 1.]
[1. 0. 0. 0. 1.]
[1. 0. 0. 0. 1.]
[1. 1. 1. 1. 1.]]Sample solution 3
import numpy as np
x = np.random.random((5,5))
print("Original Array:")
print(x)
xmin, xmax = x.min(), x.max()
print("Minimum and Maximum Values:")
print(xmin, xmax)
Original Array:
[[0.96108746 0.66272609 0.42670443 0.5463886 0.13583066]
[0.78215537 0.29811248 0.26380749 0.93835562 0.00176633]
[0.09763454 0.52875892 0.71247172 0.7065642 0.76375718]
[0.7768379 0.85771292 0.17135697 0.83372476 0.83760277]
[0.20412887 0.9024384 0.68057959 0.38648805 0.62763643]]
Minimum and Maximum Values:
0.0017663346508800526 0.9610874575010275Sample solution 4
import pandas as pd
import os
os.chdir('Whatever directory you\'ve saved cereal.csv in')
df = pd.read_csv('cereal.csv')
for index, row in df.iterrows():
print(row['name'], row['calories'])
100% Bran 70
100% Natural Bran 120
All-Bran 70
All-Bran with Extra Fiber 50
Almond Delight 110
Apple Cinnamon Cheerios 110
Apple Jacks 110
(...)
Thank you for your attention!
We will send you a survey for feedback; please take 2 minutes to answer, it helps us a lot!
